13.2 Extracting posterior samples
The function brms::posterior_samples extracts the samples from the posterior which are part of the brmsfit object.60
post_samples_temperature <- brms::posterior_samples(fit_temperature) %>% select(-lp__,-lprior)
head(post_samples_temperature)## b_Intercept b_year sigma
## 1 -4.328078 0.006692733 0.4100904
## 2 -4.202760 0.006633079 0.4050540
## 3 -2.887432 0.005926658 0.4006999
## 4 -4.580320 0.006821945 0.3996143
## 5 -2.587499 0.005782539 0.3916289
## 6 -4.247891 0.006654168 0.4081150These extracted samples can be used as before, e.g., to compute our own summary tibble:
map_dfr(post_samples_temperature, aida::summarize_sample_vector) %>%
mutate(Parameter = colnames(post_samples_temperature[1:3]))## # A tibble: 3 × 4
## Parameter `|95%` mean `95%|`
## <chr> <dbl> <dbl> <dbl>
## 1 b_Intercept -4.69 -3.51 -2.32
## 2 b_year 0.00563 0.00627 0.00689
## 3 sigma 0.373 0.405 0.440Or for manual plotting:61
post_samples_temperature %>%
pivot_longer(cols = everything()) %>%
ggplot(aes(x = value)) +
geom_density() +
facet_wrap(~name, scales = "free")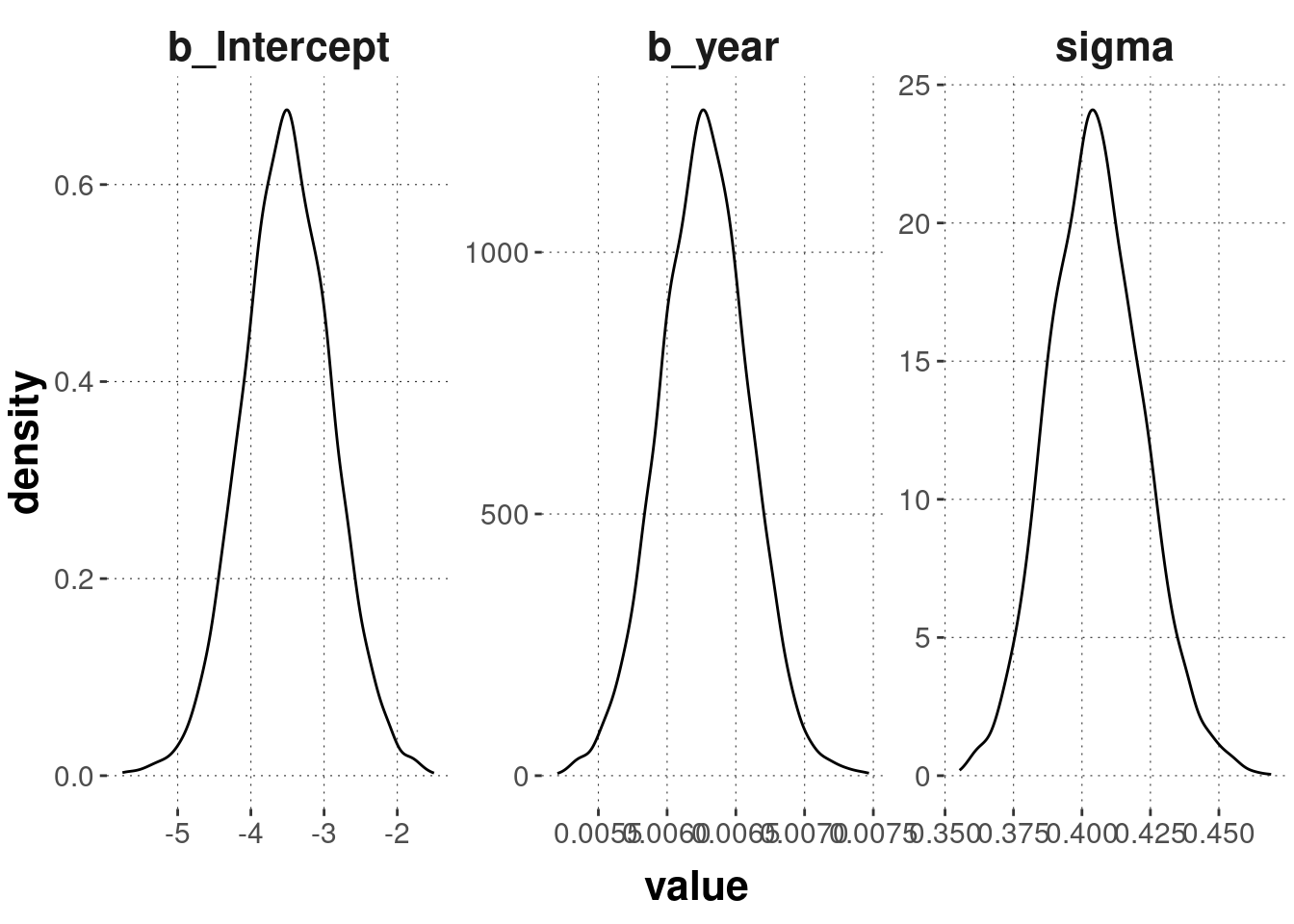
The column
lp__gives the log probability of the data for the corresponding parameter values in each row. This is useful information for model checking and model comparison, but we will neglect it here.↩︎There are also specialized packages for plotting the output of Stan models and
brmsmodel fits, such as the excellenttidybayesandggdistpackages.↩︎