Rows: 2,052
Columns: 16
$ X1 <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16…
$ trial_id <chr> "id0001", "id0002", "id0003", "id0004", "id0005", "id…
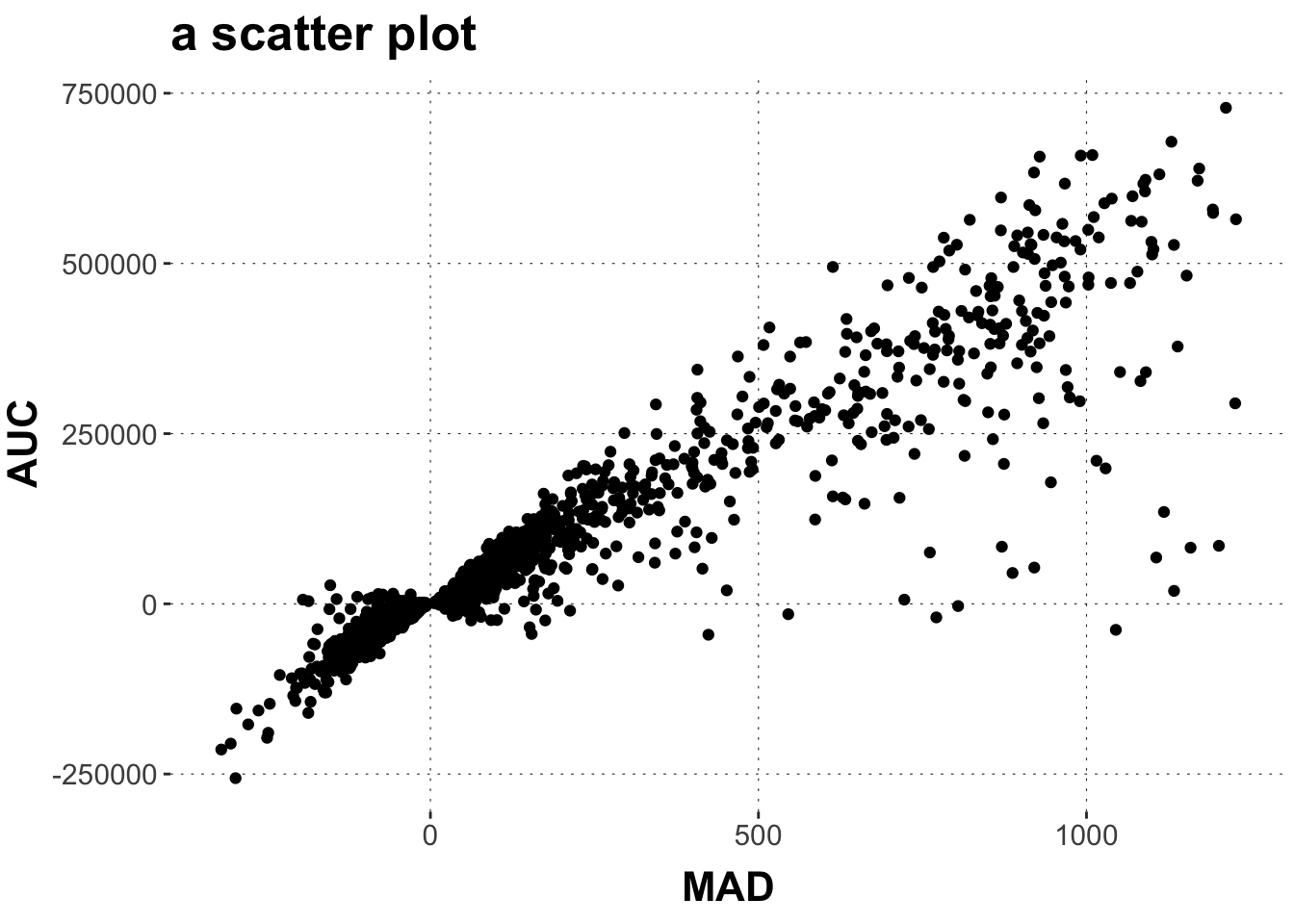
$ MAD <dbl> 82.53319, 44.73484, 283.48207, 138.94863, 401.93988, …
$ AUC <dbl> 40169.5, 13947.0, 84491.5, 74084.0, 223083.0, 308376.…
$ xpos_flips <dbl> 3, 1, 2, 0, 2, 2, 1, 0, 2, 0, 2, 2, 0, 0, 3, 1, 0, 1,…
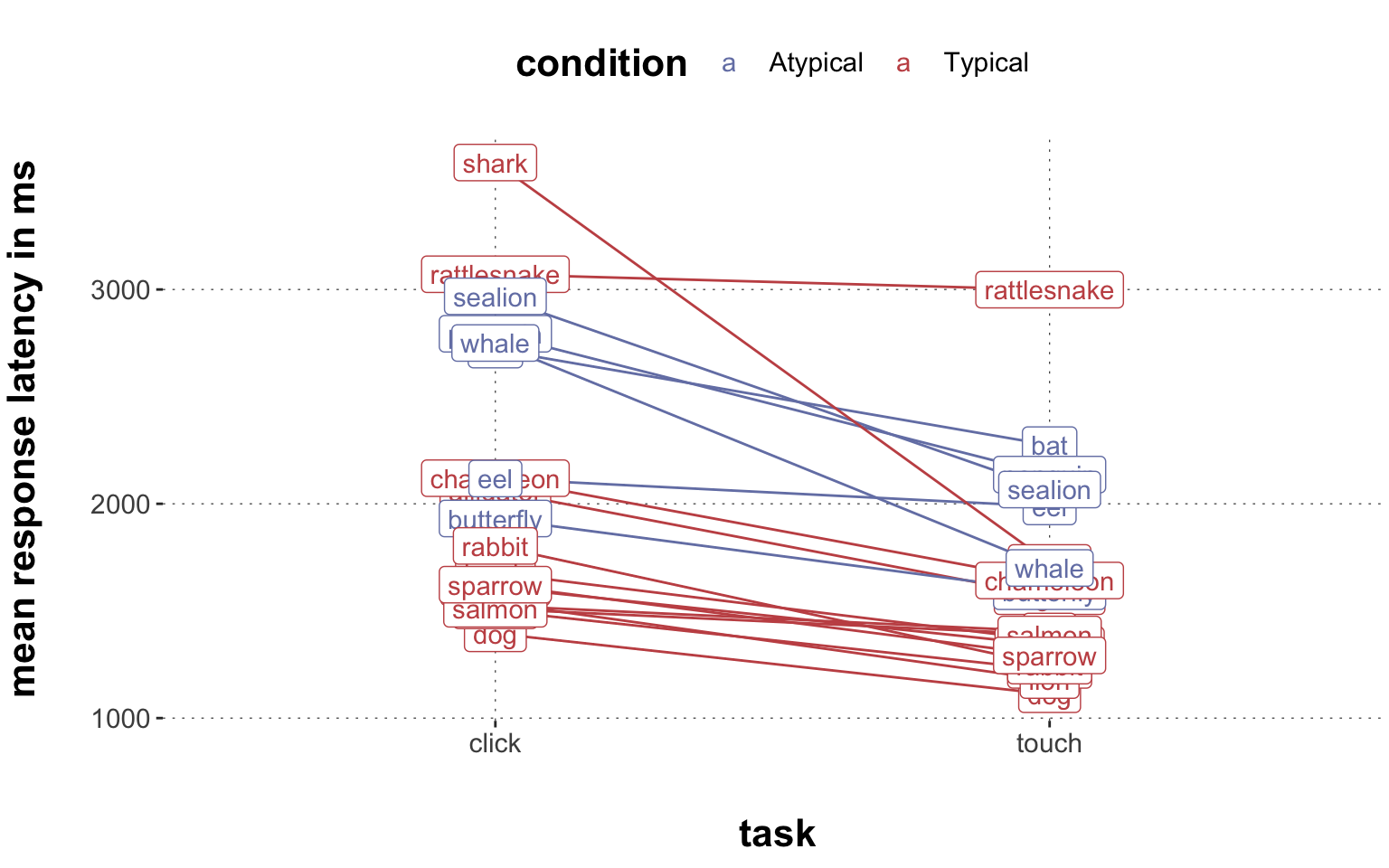
$ RT <dbl> 950, 1251, 930, 690, 951, 1079, 1050, 830, 700, 810, …
$ prototype_label <chr> "straight", "straight", "curved", "curved", "cCoM", "…
$ subject_id <dbl> 1001, 1001, 1001, 1001, 1001, 1001, 1001, 1001, 1001,…
$ group <chr> "touch", "touch", "touch", "touch", "touch", "touch",…
$ condition <chr> "Atypical", "Typical", "Atypical", "Atypical", "Typic…
$ exemplar <chr> "eel", "rattlesnake", "bat", "butterfly", "hawk", "pe…
$ category_left <chr> "fish", "amphibian", "bird", "Insekt", "bird", "fish"…
$ category_right <chr> "reptile", "reptile", "mammal", "bird", "reptile", "b…
$ category_correct <chr> "fish", "reptile", "mammal", "Insekt", "bird", "bird"…
$ response <chr> "fish", "reptile", "bird", "Insekt", "bird", "bird", …
$ correct <dbl> 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1,…