theory
posterior inference & credible values
Bayes factors & model comparison
comparison of Bayesian and "classical" NHST
practice
basics of MCMC sampling
tools for BDA ( JAGS, Stan, WebPPL, Jasp, rstanarm)
Bayesian cognitive modeling example
posterior inference & credible values
Bayes factors & model comparison
comparison of Bayesian and "classical" NHST
basics of MCMC sampling
tools for BDA ( JAGS, Stan, WebPPL, Jasp, rstanarm)
Bayesian cognitive modeling example
\[\underbrace{P(\theta \, | \, D)}_{posterior} \propto \underbrace{P(\theta)}_{prior} \times \underbrace{P(D \, | \, \theta)}_{likelihood}\]
normalizing constant:
\[ \int P(\theta') \times P(D \mid \theta') \, \text{d}\theta' = P(D) \]
how to calculate for unwieldy models with high-dimensional \(\theta\)?
an efficient way of getting samples \(x \sim P\) without access to \(P\) itself
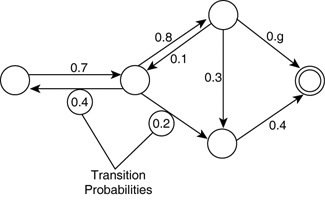
\[P(X_{n+1} = x_{n+1} \mid X_0 = x_0, \dots, X_n = x_n) = P(X_{n+1} = x_n \mid X_n = x_n)\]
dummy
good
bad

dummy dummy dummy
potential

kinetic

dummy
dummy
\(\hat{R}\)-statistics
\[\text{ESS} = \frac{N}{ 1 + 2 \sum_{k=1}^{\infty} ACF(k)}\]
| JAGS | Stan | WebPPL | |
|---|---|---|---|
| flavor | declarative | imperative | functional |
| feels like | BUGS + R | BUGS + C | JavaScript |
| methods | MH, Gibbs, … | HMC, variational, … | MH, MHM, rejection, … |
| interfaces | R, Python, Matlab | R, Python, Matlab, … | R, (…?) |
| goodies | rstanarm, loo | browser-based | |
| developers | Martyn Plummer | Stan Dev. Team (Gelman, Carpenter, …) | N. Goodman & A. Stuhlmüller et al. |
| websites | JAGS | Stan | WebPPL |
require('rjags')
modelString = "
model{
theta ~ dbeta(1,1)
k ~ dbinom(theta, N)
}"
# prepare for JAGS
dataList = list(k = 14, N = 20)
# set up and run model
jagsModel = jags.model(file = textConnection(modelString),
data = dataList,
n.chains = 2)
update(jagsModel, n.iter = 5000)
codaSamples = coda.samples(jagsModel,
variable.names = c("theta"),
n.iter = 5000)
require('ggmcmc')
require('dplyr')
ms = ggs(codaSamples)
ms %>% group_by(Parameter) %>%
summarise(mean = mean(value),
HDIlow = HPDinterval(as.mcmc(value))[1],
HDIhigh = HPDinterval(as.mcmc(value))[2])
## # A tibble: 1 × 4 ## Parameter mean HDIlow HDIhigh ## <fctr> <dbl> <dbl> <dbl> ## 1 theta 0.6814352 0.4967396 0.8756536
tracePlot = ggs_traceplot(ms)
densPlot = ggs_density(ms) +
stat_function(fun = function(x) dbeta(x, 15,7), color = "black")
require('gridExtra')
grid.arrange(tracePlot, densPlot, ncol = 2)
Your turn: what's this model good for?
obs is a vector of 100 observations (real numbers)model{
mu ~ dunif(-2,2)
var ~ dunif(0, 100)
tau = 1/var
for (i in 1:100){
obs[i] ~ dnorm(mu,tau)
}
}
NB: JAGS' precision \(\tau = 1/\sigma^2\), not standard deviation \(\sigma\) in dnorm
model{
mu ~ dunif(-2,2)
var ~ dunif(0, 100)
tau = 1/var
for (i in 1:100){
obs[i] ~ dnorm(mu,tau)
}
}
\[\mu \sim \text{Unif}(-2,2)\] \[\sigma^2 \sim \text{Unif}(0,100)\] \[obs_i \sim \text{Norm}(\mu, \sigma)\]
actively developed by Andrew Gelman, Bob Carpenter and others
uses HMC and NUTS, but can do variational Bayes and MAP too
written in C++
cross-platform
interfaces: command line, Python, Julia, R, Stata, Matlab
require('rjags')
ms = "
model{
theta ~ dbeta(1,1)
k ~ dbinom(theta, N)
}"
# prepare data for JAGS
dataList = list(k = 14, N = 20)
# set up and run model
jagsModel = jags.model(
file = textConnection(ms),
data = dataList, n.chains = 2)
update(jagsModel, n.iter = 5000)
codaSamples = coda.samples(
jagsModel,
variable.names = c("theta"),
n.iter = 5000)require('rstan')
ms = "
data {
int<lower=0> N ;
int<lower=0> k ;
}
parameters {
real<lower=0,upper=1> theta ;
}
model {
theta ~ beta(1,1) ;
k ~ binomial(N, theta) ;
}"
# prepare data for Stan
dataList = list(k = 14, N = 20)
# set up and run
mDso = stan_model(model_code = ms)
stanFit = sampling(object = mDso,
data = dataList,
chains = 2,
iter = 10000,
warmup = 5000)plot(stanFit)
codaSamples = As.mcmc.list(stanFit) ms = ggs(stanFit) ggs_density(ms)
dummy
15 people answered 40 exam questions. Here's their scores:
k <- c(21,17,21,18,22,31,31,34,34,35,35,36,39,36,35) p <- length(k) #number of people n <- 40 # number of questions
dummy dummy dummy dummy dummy dummy
[from Lee & Wagenmakers (2015), Chapter 6.1]
model{
for (i in 1:p){
z[i] ~ dbern(0.5)
}
psi = 0.5
phi ~ dunif(0.5,1)
for (i in 1:p){
theta[i] = ifelse(z[i]==0,psi,phi)
k[i] ~ dbin(theta[i],n)
}
}k <- c(21,17,21,18,22,31,31,34,34,35,35,36,39,36,35) show(summary(codaSamples)[[1]][,1:2])
## Mean SD ## phi 0.863443 0.01719438 ## z[1] 0.000000 0.00000000 ## z[2] 0.000000 0.00000000 ## z[3] 0.000000 0.00000000 ## z[4] 0.000000 0.00000000 ## z[5] 0.000000 0.00000000 ## z[6] 0.992200 0.08797689 ## z[7] 0.994500 0.07396146 ## z[8] 1.000000 0.00000000 ## z[9] 0.999900 0.01000000 ## z[10] 1.000000 0.00000000 ## z[11] 1.000000 0.00000000 ## z[12] 1.000000 0.00000000 ## z[13] 1.000000 0.00000000 ## z[14] 1.000000 0.00000000 ## z[15] 1.000000 0.00000000
data {
int<lower=1> p; int<lower=0> k[p]; int<lower=1> n;
}
transformed data {
real psi; psi <- .5;
}
parameters {
real<lower=.5,upper=1> phi;
}
transformed parameters {
vector[2] log_alpha[p];
for (i in 1:p) {
log_alpha[i,1] <- log(.5) + binomial_log(k[i], n, phi);
log_alpha[i,2] <- log(.5) + binomial_log(k[i], n, psi);
}
}
model {
for (i in 1:p)
increment_log_prob(log_sum_exp(log_alpha[i]));
}
generated quantities {
int<lower=0,upper=1> z[p];
for (i in 1:p) {
z[i] <- bernoulli_rng(softmax(log_alpha[i])[1]);
}
}
\[\log P(k_i, n, z_i, \phi) = \log \text{Bern}(z_i \mid 0.5) + \log \text{Binom}(k_i, n, \text{ifelse}(z=1, \phi, 0.5))\]
\[ \begin{align*} P(k_i, n, \phi) & = \sum_{z_i=0}^1 \text{Bern}(z_i \mid 0.5) \ \text{Binom}(k_i, n, \text{ifelse}(z=1, \phi, 0.5)) \\ & = \text{Bern}(0 \mid 0.5) \ \text{Binom}(k_i, n, 0.5)) + \\ & \ \ \ \ \ \ \ \ \ \ \text{Bern}(1 \mid 0.5) \ \text{Binom}(k_i, n, 1)) \\ & = \underbrace{0.5 \ \text{Binom}(k_i, n, 0.5))}_{\alpha_0} + \underbrace{0.5 \ \text{Binom}(k_i, n, 1))}_{\alpha_1} \\ \log P(k_i, n, \phi) & = \log \sum_{i=0}^1 \exp \log \alpha_i \end{align*}\]
library(rwebppl)
datainput = list(n = 24, k = 7)
modelString = '
var successes = dataFromR["k"][0];
var trials = dataFromR["n"][0];
var model2compute = function(){
var p = uniform(0,1);
observe(Binomial({n: trials, p: p}), successes);
return {bias: p}; }'
wp.rs = webppl(modelString, model_var = "model2compute",
data = datainput,
data_var = "dataFromR",
inference_opts = list(method = "MCMC",
samples = 15000,
burn = 5000, verbose = TRUE),
output_format = "ggmcmc",
chains = 2, cores = 2)
DEMO
- liberate psychology from expensive, proprietory software (SPSS) - provide Bayesian statistical methods in an accessible way - provide a universal platform for publishing statistical analysis with rich UIs
DEMO
require('datasets')
head(cars)
m1 = glm(speed ~ dist, data = cars)require('rstanarm')
m2 = stan_glm(speed ~ dist, data = cars)[check this vignette, and this great tutorial paper for more]